Let’s discuss the question: pymol show hydrogen bonds. We summarize all relevant answers in section Q&A of website Myyachtguardian.com in category: Blog MMO. See more related questions in the comments below.

Table of Contents
Can you show bonds in PyMOL?
Bonds. PyMOL can deduce bonds from the PDB structure file, even if the CONECT records are missing.
How do you show interactions in PyMOL?
The easiest way to observe any receptor-ligand interaction in PyMOL is! (1) Load your complex. (2) Hide water etc if any for visibility enhancement… (3) Click the action button (A) and scroll down to presets that will walk you through to another list where you will find “ligand sites”.
Pymol for Beginners – video 4: H-bonds
Images related to the topicPymol for Beginners – video 4: H-bonds

How do you visualize hydrogen bonds in VMD?
- Go to the Extension–>Analysis–> Hydrogen Bonds.
- In the Hydrogen bond tab, you will see, Selection 1, assign there protein, and in selection 2 assign the residue name or residue id of your ligand.
How do you show double bonds in PyMOL?
Turning on the valence setting will enable the display of double bonds. valence_size alters the distance of double bonds. Note that bonds can be edited to be delocalized using Unbond and Bond.
How do you show disulfide bonds in PyMOL?
Pymol has a special function for showing disulfide bonds. To do so, go to the “S‟ menu then put the cursor on “disulfides. ‟ Display the disulfide in the representation of your choice.
What is the length of a typical hydrogen bond?
Hydrogen Bonding
If a proper hydrogen bond acceptor–donor pair is within the correct distance, the bond is taken to be a hydrogen bond. This distance is generally considered to be from 2.7 to 3.3 Å, with 3.0 Å being the most common value for protein and water hydrogen bonds.
How do I add a bond in PyMOL?
You can easily create a new bond by selecting two atoms, each with the CTRL-MIDDLE-MOUSE-BUTTON and typing “bond” on the command line.
How do you identify a hydrogen bond donor and acceptor?
The donor in a hydrogen bond is usually a strongly electronegative atom such as N, O, or F that is covalently bonded to a hydrogen bond. The hydrogen acceptor is an electronegative atom of a neighboring molecule or ion that contains a lone pair that participates in the hydrogen bond.
PyMol Measuring Distance with Hydrogen Bonding Interactions 2020 11 11
Images related to the topicPyMol Measuring Distance with Hydrogen Bonding Interactions 2020 11 11

Does H2S have hydrogen bonding?
For example, consider hydrogen sulfide, H2S, a molecule that has the same shape as water but does not contain hydrogen bonds. Due to its relatively weak intermolecular forces, H2S boils at about −60 °C and so is a gas at room temperature. In contrast, water boils at 100 °C.
How do you cite a Ligplot?
Wallace A C, Laskowski R A & Thornton J M (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Prot. Eng., 8, 127-134.
How do you do protein ligand docking?
- 3.1. Target selection. …
- 3.2. Ligand selection and preparation. …
- 3.3. Docking. …
- 3.4. Evaluating docking results. …
- 3.5. Docking software description.
How do you determine the number of hydrogen bond donors?
Donor count = the sum of the atoms in the molecule which have H donor property. Donor sites = the sum of the H atoms connected to the donor atoms. Acceptor count = the sum of the acceptor atoms. An acceptor atom always has a lone electron pair/lone electron pairs that is capable of establishing a H bond.
How do I use VMD?
- In the OpenGL Display, press the left mouse button down and move the mouse. …
- Holding down the right mouse button and repeating the previous step will cause rotation around an axis perpendicular to the screen (Figure 3(b)).
How do you show ball and stick in Pymol?
Using the mouse, for the desired object or selection click A > Preset > ball and stick.
How do you detect a disulfide bond?
Researchers have successfully demonstrated that disulfide bridge patterns can be identified by mas spectrometry (MS) analysis, following protein digestion either under partial reduction12,13,16,17 or nonreduction conditions. Partial reduction is a widely accepted approach for the determination of disulfide bonds.
Analysing Protein-Ligand Interactions : Tutorial
Images related to the topicAnalysing Protein-Ligand Interactions : Tutorial
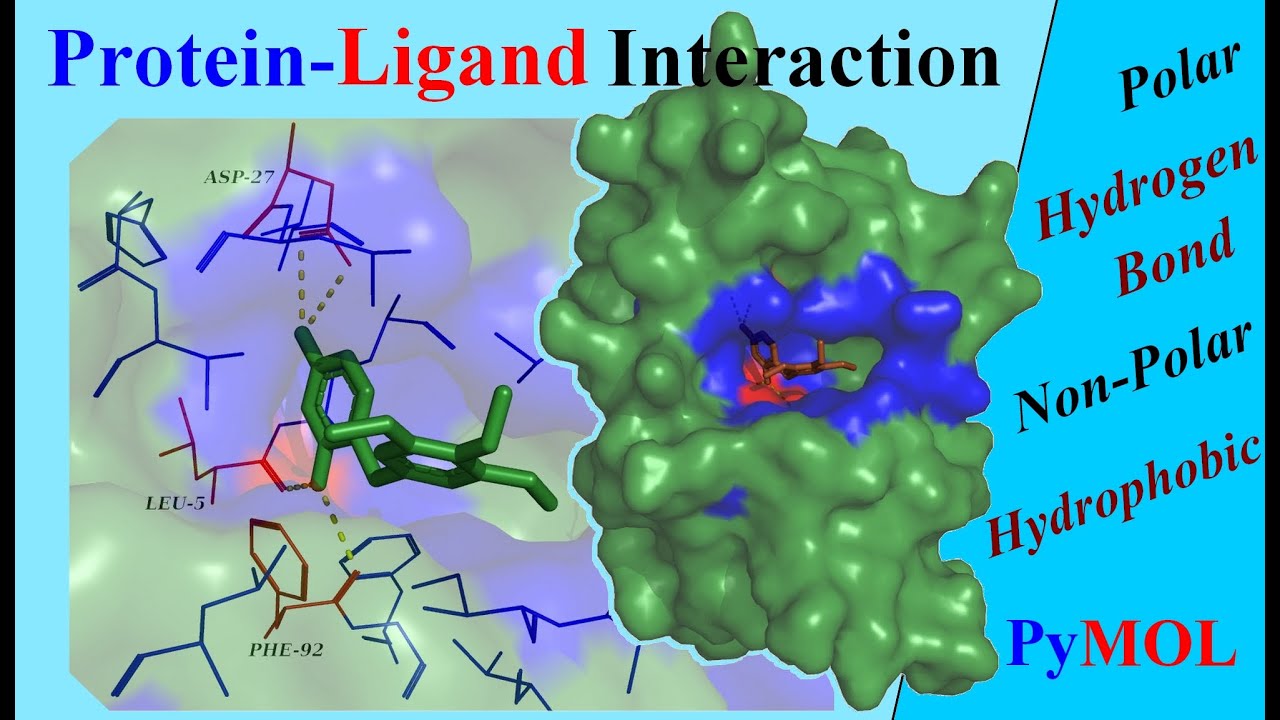
How do you tell if a protein has a disulfide bond?
Disulfide cross-linkages may be located by cleaving a protein between half-cystinyl residues to give peptides that contain only one disulfide bond. The molecular weights of these peptides are determined by fast atom bombardment mass spectrometry (FAB-MS) and related to specific segments of the parent protein.
How do you see the disulfide bonds in a chimera?
Enable the display of disulfide bonds with “Select->Residue->CYS” followed by “Actions->Atoms/Bonds->Show”. (Once the cysteines are displayed, you can deselect them by ctrl-clicking on the background or by clicking “Select- >Clear Selection”.)
Related searches
- how to show hydrogen bonds
- pymol create bond between objects
- pymol show hydrogen atoms
- pymol hydrogen bond distance
- how to calculate hydrogen bonds
- pymol find any contacts
- show h bonds in pymol
- hide hydrogens pymol
- pymol select cofactor
- how to show hydrogen bonds in chimera
- pymol hydrogen bond thickness
- how does hydrogen bonds form
- how many pi bonds can sp3 form
- pymol select
- how to find disulfide bonds in pymol
- how many sigma bonds in sp3
- how many sigma and pi bonds in sp3
Information related to the topic pymol show hydrogen bonds
Here are the search results of the thread pymol show hydrogen bonds from Bing. You can read more if you want.
You have just come across an article on the topic pymol show hydrogen bonds. If you found this article useful, please share it. Thank you very much.

